Compact Modeling Techniques for Enabling Full-Body Cardiovascular Simulation
- Category: Medical Electronics
- Tags: luca daniel, yu-chung hsiao
The recent development of cardiovascular simulation furthers the understanding of physiological conditions of blood-related disorders. For example, detailed finite volume simulation of aorta segments has revealed the susceptible sites for atherosclerotic lesion formation[1]. The simulated interaction between a thrombus and an installed intravenous filter in a single vena cava described in[2] reflects the concerns of device migration and failure due to the change of velocity and pressure profile. When performing simulation at the “full-body” level, the state-of-the-art techniques rely on significant approximations of components of the circulatory network to circumvent formidable complexity. For instance, organs and capillaries are simplified as lumped pressure resistance; vessels are manually reduced to one or two-dimensions; Navier-Stokes equations are linearized; and the elasticity of vessel walls is selectively neglected[3]. The aforementioned approximations are typically performed empirically. Therefore, the aggregate loss of accuracy and the missing nonlinear behaviors at the full-body level are difficult to predict.
In this project, we are developing algorithms that can automatically construct nonlinear dynamical models from terminal pressure-flow relationships and guarantee that the models are accurate, compact, and suitable for network composition[4]). We use physics-specific field solvers that do not compromise on accuracy to construct elaborate local models for circulatory parts, such as porous types of organs, tree-like capillaries, and general vessel bifurcation where turbulent flows may occur (Figure 1). Our algorithms then extract efficient and compact models. Such an approach enables large-scale network modeling that is computationally feasible and able to capture the previously neglected nonlinear phenomena. When we search for optimal model coefficients, our algorithms ensure that the resulting models do not generate energy, which in turn implies that a composed network is also dissipative. Hence the full-body network can be composed bottom-up and behave stably in time-domain simulations.
- Figure 1: Left: Pressure profile of a vessel bifurcation solved by finite element method. Length of each branch is chosen such that Poiseuille flow is fully developed to satisfy network port assumptions. Right: Compact model extracted by our algorithms. Models can be further parameterized with vessel geometries.
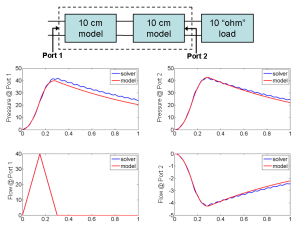
- Figure 2: A 20-cm artery is constructed by cascading two 10-cm artery models terminated with a matched load. The transient simulation results of the cascaded model are compared with those of a 20-cm artery solved by our immersed boundary method solver.
- J. R. Buchanan, C. Kleinstreuer, S. Hyun, and G. A. Truskey, “Hemodynamics simulation and identification of susceptible sites of atherosclerotic lesion formation in a model abdominal aorta,” Journal of Biomechanics, vol. 36, issue 8, pp. 1185-1196, Aug. 2003. [↩]
- Y. V. Vassilevski, S. S. Simakov, and S. A. Kapranov, “A multi-model approach to intravenous filter optimization,” International Journal for Numerical Methods in Biomedical Engineering, vol. 26, issue 7, pp. 915-925, July 2010. [↩]
- A. Quarteroni, M. Tuveri, and A. Veneziani “Computational vascular fluid dynamics: problems, models and methods,” Computing and Visualization in Science, vol. 2, no. 4, pp. 163-97, 2000. [↩]
- B. Bond, T. Moselhy, and L. Daniel, “System identification techniques for modeling of the human arterial system,” in Proc. SIAM Conference on the Life Sciences, Pittsburgh, PA, July 2010, p. 12-15. (invited [↩]