- Authors: J. B. Chang, H. K. Choi, A. F. Hannon, C. A. Ross, K. K. Berggren
- Sponsorship: TSMC, SRC
Topographic templates can be used for guiding the self-assembly of block copolymers to produce complex nanoscale patterns. In our previous work, sub-20-nm bends or meander structures were achieved based on topographic templates using 45.5 kg/mol polystyrene polydimethylsiloxane (PS-PDMS) block copolymer [] . To template the self-assembly of 16 kg/mol PS-b-PDMS with a sub-10-nm periodicity, so as to scale down the resolution to sub-10-nm, the diameter of the templates must also be scaled down to the sub-10-nm range. However, the fabrication of sub-10-nm posts is challenging even with state-of-the-art lithography.
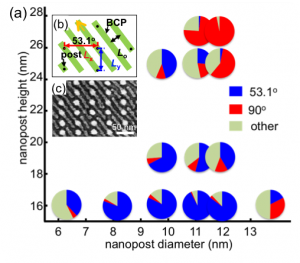
In this study, we demonstrate aligned sub-10-nm block copolymer patterns, based on the understanding of the interaction between the block copolymer cylinders and posts with a range of diameters and heights. Figure 1 shows how the orientation of the block copolymer cylinders depended on the post diameter for a given spacing, Lx = 48 nm and Ly = 32 nm. The commensurate orientation of the 16 kg/mol PS-b-PDMS block copolymer cylinders for this array period is depicted in Figure 1(b), giving a cylinder orientation of 53.1˚ with respect to the x-axis. The area fraction of the block copolymer oriented along the commensurate orientation was over >75% when templated by posts with a height of 16 nm and diameters from 8 nm to 12 nm. The cylinders were not aligned in a preferential orientation when templated by posts with a diameter of 6 nm. However, the cylinders were aligned parallel to the y-axis when templated by large (diameter > 13 nm) or tall (height > 24 nm) posts.
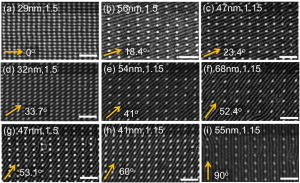
Based on the preceding result, we demonstrated every possible commensurate condition for the 18-nm period PS-b-PDMS. As shown in Figure 2, the orientation of the block copolymer cylinders could be varied between 0o and 90o with respect to the x-axis by using posts with a height of 19 nm and a diameter of 10 nm.
-
-
Figure 1: (a) The change of area fraction of the block copolymer cylinders templated by a post array with Lx = 48 nm and Ly = 32 nm depending on the post diameter and height. (b) The commensurate orientation for a post array with Lx = 48 nm and Ly = 32 nm. (c) SEM image of the PDMS cylinders templated by the post array with a post diameter of 12 nm and a post height of 16 nm.
-
-
Figure 2: SEM images of PDMS cylinders templated by post arrays. The height of the posts was 19 nm and the diameter was 10 nm. Arrows indicate the direction of the PDMS cylinders. Each figure is annotated Lx, Lx/Ly. Scale bar = 100 nm.
- Authors: Y. Kim, K. E. Thompson, J. B. Chang, V. R. Manfrinato, A. E. Keating, K. K. Berggren
- Sponsorship: International Iberian Nanotechnology Laboratory
Templated assembly of biomolecules can create complex nanostructured devices with precisely tailored chemical or biological responses, with applications in, for example, nanoscale patterning for electronics, biomedical devices, or environmental sensors. In this research project, we focus on developing methods for creating complex molecular top-down templating of assembled structures of protein that will be relevant to a range of devices.
We examined a range of EM staining techniques for cortexillin, which is coiled-coil protein and forms a parallel homodimer as an actin-binding domain. The protein constructs have the addition of single cysteine residues at either the N- or C- terminus that would facilitate binding gold metal surfaces, as Figure 1(A) shows. Weakly staining the rod shape of proteins in uranyl acetate resulted in observation of structures 20 nm in length and 4~5 nm in width, which roughly matches the expected protein model from crystallographic structure, as in Figure 1(B). We also tested tagging of the cortexillin homodimer using gold nanoparticles, which resulted in gold points colocalized to the proteins. This provides a straightforward way to visualize single protein molecules using TEM and SEM.
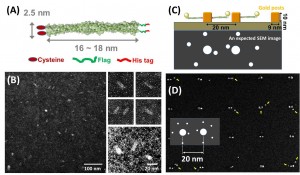
Figure 1: (A) The structure of cortexillin (B) TEM images of cortexillin stained with 2% uranyl acetate and inset images (right) showing morphology of protein such as fat rods in higher magnification. (C) Single molecular protein array in the project, presents gold tagging to His-tag in protein and Cys residue facilitates binding to gold posts. (D) SEM images of protein array in the gold post pattern with 20-nm pitch.
We are also been developing a method to position proteins into the patterned surfaces with sub-10-nm resolution. We have functionalized an e-beam patterned surface with gold, resulting in two-post arrays with a pitch varying from 10 to 50 nm. The cysteine of the cortexillin homodimer can then be used to direct the attachment of the protein to the posts via thiol-gold attachment chemistry. The cortexillin proteins also contain a C-terminal hexa-histidine tag, which allows for the attachment of Ni-NTA gold nanoparticles (NTA: nitrilotriacetic acid) via the coordination of the charged histidines around the nickel ion (Kd ≅ 10-6 M). Figure 1(C) illustrates the assembly scheme of single protein array on a gold post pattern, where the cysteine residue in a coiled-coil protein that allows binding to surfaces of gold posts and the hexa-histidine sequence allows attachment of Ni-NTA gold nanoparticles. By performing multiple step protocol of: e-beam patterning of poly methyl methacrylate (PMMA) resist, gold post generation on PMMA pattern, PMMA development, cleaning by Plasma etching, and incubation in the Cys-modified protein solution, we achieved results suggesting the formation of single-molecule protein arrays around the gold posts of the pattern as observed by SEM. SEM imaging of the pattern after incubation with protein reveals that there is a relatively low density of protein binding, as indicated by small gold nanoparticles around the gold post patterns, as Figure 1(D) shows. Here, we could develop the method using electron-beam lithography to create a gold post pattern with sub-10-nm resolution pitch close to biomolecular scale, which could then be used to template novel systems including DNA, protein, and other biomolecules.


