Cell Sorting and Identification for Immunology
- Category: MEMS & BioMEMS
- Tags: joel voldman, marc castelarnau
A major challenge in immunobiology is to better understand how the immune cells dynamically interact with each other and with their environment. Any insights in that direction can help provide a clearer picture of the immune response’s evolution and reaction to infectious diseases; to this end, the use of clinical samples is extremely valuable [1] . However, most of the current analytical techniques used to characterize cells in a sample (e.g., ELISA, flow cytometry, PCR, DNA/RNA sequencing) do not preserve the cells after the analysis, and they typically fail to give multiple parameters of interest from the same sample. In addition, those analyses return only a snapshot of the sample state at a given time, which in most cases cannot be realistically compared with the in vivo conditions. Therefore, there is a need to develop new approaches for high-throughput and multiparameter analyses on clinical samples in a time-dependent manner and with single-cell resolution, to obtain the maximum information from a clinical sample.
Recently developed single-cell, multiparameter analytical platforms [2] [3] are oriented to this purpose and can be complemented with DNA/RNA analysis and sequencing to provide a complete picture of the immune cells from a sample, as illustrated in Figure 1. Nevertheless, linking the results of those different approaches requires keeping the identity of the cells all along the multiple analyses. The proposed solution consists of four steps (i) a stochastic bead-based labeling of the cells of interest in the multiparameter analytical platform, (ii) imaging of the labeled cells, (iii) an optical cell release to allow sorting of the selected cells [4] and finally, (iv) a read-out of the labels on the cells in the final platform to re-assign the cell identity.
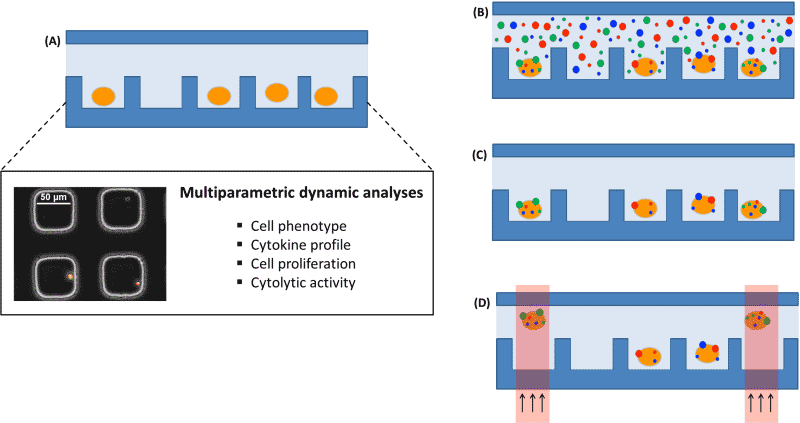
Figure 1: Schematic of the envisioned multiparameter analytical platform. (A) Cells settle down in a microwell array for multiparameter dynamic analysis. (B) Cells randomly labeled with beads. (C) Imaging of the labels on selected cells. (D) Optical-based sorting of the selected cells for genetic analysis.
- T. D. Querec, R. S. Akondy, E. K. Lee, W. Cao, H. I. Nakaya, D. Teuwen, A. Pirani, K. Gernert, J. Deng, B. Marzolf, K. Kennedy, H. Wu, S. Bennouna, H. Oluoch, J. Miller, R. Z. Vencio, M. Mulligan, A. Aderem, R. Ahmed, and B. Pulendran, “Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans,” Nature Immunology, vol. 10, pp. 116-125, 2008. [↩]
- J. C. Love, J. L. Ronan, G. M. Grotenbreg, A. G. van der Veen, and H. L. Ploegh, “A microengraving method for rapid selection of single cells producing antigen-specific antibodies,” Nature Biotechnology, vol. 24, pp. 703-707, 2006. [↩]
- J. C. Love, “Integrated process design for single-cell analytical technologies,” AIChE Journal, vol. 56, pp. 2496-2502, 2010. [↩]
- J. R. Kovac and J. Voldman, “Intuitive, Image-Based Cell Sorting Using Optofluidic Cell Sorting,” Analytical Chemistry, vol. 79, pp. 9321-9330, 2007. [↩]