Micropatterning of Cells to Study Autocrine Signaling
- Category: MEMS & BioMEMS
- Tags: Joel Voldman, Somponnat Sampattavanich
Autocrine signaling is a mode of chemical signaling that occurs when cells can capture self-secreted diffusive factors. Apart from its major role in sustaining cancer growth, autocrine signaling is also involved in the positive-feedback regulation of various physiological processes. Due to the closed-loop nature and complex interplay of this signaling with other signaling cues, it is difficult to validate the presence and function of autocrine loops. Studying these loops typically requires the use of specific inhibitors to perturb the underlying ligand/receptor pairing, limiting investigation of poorly characterized autocrine loops.
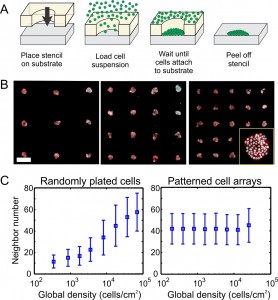
To promote the examination of autocrine signaling in broader biological systems, we have developed a general method for modulating autocrine activity using cell patterning. In addition to capturing self-secreted ligands, cells with autocrine loops also acquire ligands from their neighbors. By modulating the relative positioning between cells, we are able to modulate capture of autocrine ligands without needing specific inhibitors. In particular, we use stencil cell patterning to organize cells as square-latticed arrays of circular patches of varying array spacing (Figures 1A & B). We found that the cell-patterning platform can maintain uniform local cell density at all array spacings, in contrast to randomly plated cells, which exhibit increasing local cell density (Figure 1C). By reducing the influence of these other environmental cues, we are able to more explicitly study the effect of autocrine signaling on cell phenotype.
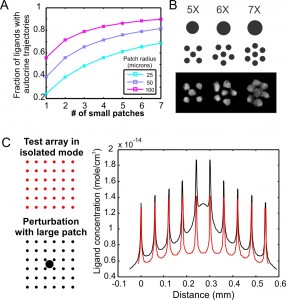
In addition to studying the direct role of autocrine signaling, the cell-patterning platform can also be used to investigate the interplay of autocrine signaling with other signaling cues and to evaluate its contribution towards cell-to-cell variability. To determine the concurrent role of cell-cell contacts, we can compare cell responses between single patches and multiple patches where the cell number of both designs is equal (Figures 2A & B). To evaluate the contribution of autocrine loops in causing cell-to-cell variability, we can determine how the inclusion of a large cell patch will perturb the response of an array of small patches (Figure 2C). These innovative cell-patterning designs provide us novel tools for characterizing the impact of autocrine signaling without prior knowledge of the underlying ligand/receptor interactions.
- Figure 1: Investigation of autocrine signaling using cell patterning. A) Stencil patterning was used to organize the square-latticed arrays of circular patches. B) Resulting cell patterns of the A431 epidermoid carcinoma cells of varying array spacing. Scale bar, 500 µm. C) Changes of local density (e.g., neighbor number) with alteration of global cell density in randomly plated and patterned cell arrays.
- Figure 2: Creative cell-patterning designs. A) Calculated fractions of ligands with autocrine trajectories for varying total patch areas and patch radii. B) Examples of single-patch designs with varying radii, the composite-patch designs with the same total area, and the actual resulting patterns. C) Designs of cell arrays for studying autocrine signaling contribution on cell-to-cell variability. The right-hand plot illustrates predicted ligand concentration for both designs.