Image-based Sorting of Cells
- Category: MEMS & BioMEMS
- Tags: Joel Voldman, Joseph Kovac, Tao Sun
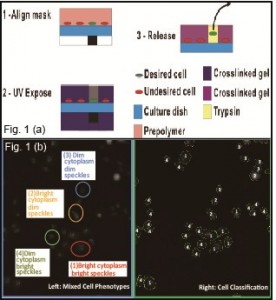
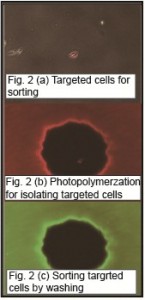
Microfabricated/microfluidic approaches to cell sorting, include purely dielectrophoretic (DEP) trap arrays [1] , passive hydrodynamic trap arrays with active DEP-based cell release [2] , and passive microwell arrays with optical cell release to permit sorting of non-adhered cells [3] . As these proceeding technologies were best suited to operate with non-adherent cells, we are developing a solution for adherent cells. Our approach to sorting adherent cells based on the morphological features uses a photolithography-inspired method, illustrated in Figure 1 (a). We first plated adherent cells into a dish and imaged cells using a microscope. Machine learning algorithm-based software CellProfiler [4] was used to quantitatively characterize the morphological features of the imaged cells, covering the cell area, shape, fluorescent intensity, and texture. As shown in Figure 1 (b), four classes were defined according to the fluorescent intensity difference in cell cellular compartments. A set of judging rules was generated by iteratively training the classifier based on hundreds of quantitative cell feature measurements to cluster the cells of similar phenotypes into a particular class. Desired cells were identified according to the classification. An alignment mark image was generated, with black features corresponding to locations of desired cells. Aligning the transparency mask to the back of the cell culture dish showed that opaque mask features resided beneath desired cells. We then mixed a prepolymer solutizon consisting of cell culture media, a UV-photoinitiator, and poly(ethylene glycol) diacrylate (PEGDA) monomer. We added the prepolymer to the cell culture dish and shined ultraviolet (UV) light from a standard fluorescence microscope fluorescence source through the transparency and into the dish. The prepolymer then crosslinked into a hydrogel in all unmasked locations, encapsulating undesired cells. The desired cells, which were not encapsulated, can be enzymatically released from the substrate and recovered, as shown in Figure 2. The overall technique requires standard equipment found in biological labs and inexpensive reagents (<$10 per experiment), encouraging widespread adoption.
- Figure 1: (a) Schematic of sorting method. (b) Image showing the automated cell classification using CellProfiler. Left: mixed cell phenotypes of four classes, Right: results of segmented individual cells and classified cell populations.
- Figure 2: (a) Image showing targeted cells (red) in the dish (b) Image showing the formation of photo-polymerized gel microwells (500-µm diameter) to isolate the targeted cells. (c) Image showing the remaining microwells after cell sorting.
- A. Rosenthal and J. Voldman, “Dielectrophoretic traps for single-particle patterning,” Biophys. J., vol.88, pp. 2193-2205, 2005. [↩]
- B. M. Taff, S. P. Desai, and J. Voldman, “Electroactive hydrodynamic weirs for micro-particle manipulation and patterning,” Applied Physics Letters, vol. 94, no. 8, p. 084102, 2009. [↩]
- J. R. Kovac and J. Voldman, “ Intuitive, image-based cell sorting using opto-fluidic cell sorting,” Analytical Chemistry, vol. 79, pp. 9321-9330, 2007. [↩]
- T. R. Jones, A. E. Carpenter, M. R. Lamprech, J. Moffat, S. J. Silver, J. K. Grenier, A. B. Castoreno, U. S. Eggert, D. E. Root, P. Golland, and D. M. Sabatini, “Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning,” PNAS, vol. 106, pp.1826-1831, 2009. [↩]