DNA-templated Assembly of Droplet-derived Microtissues
- Category: MEMS & BioMEMS
- Tags: Cheri Li, Sangeeta Bhatia
Paracrine and autocrine cell signaling are critical factors guiding tissue development and maintenance, and dysregulation of these cues contributes to the pathogenesis of diseased states such as cancer. Patterning multiple cell types is thus a critical step for engineering functional tissue [1] , but current “top-down” approaches such as dielectrophoresis and photopatterning are challenging to scale-up for the construction of mesoscale tissues. On the other hand, “bottom-up” methods wherein small tissue building blocks are assembled into larger structures have potential for constructing multicellular tissues in a facile, scalable fashion. Synthetic microtissues composed of cell-laden hydrogels in this size range represent appropriate fundamental building blocks of such bottom-up methods [2] .
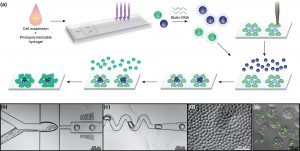
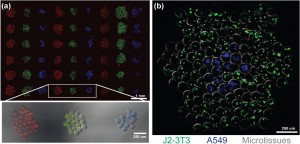
To specify the placement of many different microtissues relative to one another, we have developed a “bottom-up” approach for fabricating multicellular tissue constructs that utilizes DNA-templated assembly of 3D cell-laden hydrogel microtissues (Figure 1a). A microfluidic flow focusing-generated emulsion of photopolymerizable prepolymer is used to produce 100-µm monodisperse microtissues at a rate of 100 Hz (105/hr) (Figure 1b-d). Multiple cell types, including suspension and adherently cultured cells, can be encapsulated into the microtissues with high viability (~97%) (Figure 1e). We then use a DNA coding scheme to self-assemble microtissues “bottom-up” from a template that is defined using “top-down” techniques. The microtissues are derivatized with single-stranded DNA using a biotin-streptavidin linkage to the polymer network and are assembled by sequence-specific hybridization onto spotted DNA microarrays. Using orthogonal DNA codes, we have achieved multiplexed patterning of multiple microtissue types with high binding efficiency and >90% patterning specificity (Figure 2a). We have also demonstrated the ability to organize multicomponent constructs composed of epithelial and mesenchymal microtissues while preserving each cell type in a 3D microenvironment (Figure 2b).
- Figure 1: (a) Schematic of microtissue fabrication, functionalization, and DNA-templated self-assembly. (b) Cells and prepolymer flow into a 60-µm droplet-generating nozzle. (c) Droplets pass through a mixer and are then UV-polymerized. (d) Microtissues collected from the device. (e) Fibroblasts (J2-3T3) encapsulated and stained for viability (green-live, red-dead).
- Figure 2: (a) Three-color (RGB) microtissue assembly using orthogonal sequences. Microtissues contain marker beads. (b) Microtissues encapsulating either J2-3T3 (green) or A549 cells (blue) assembled into composite clusters.
- S. March, E. E. Hui, G. H. Underhill, S. Khetani, and S. N. Bhatia, “Microenvironmental regulation of the sinusoidal endothelial cell phenotype in vitro,” Hepatology, vol. 50, pp. 920-8, 2009. [↩]
- A. A. Chen, G. H. Underhill, and S. N. Bhatia, “Multiplexed, high-throughput analysis of 3D microtissue suspensions,” Integr Biol (Camb), vol. 2, pp. 517-27, 2010. [↩]